A new article by faculty members has been published in the journal Biochimica et Biophysica Acta
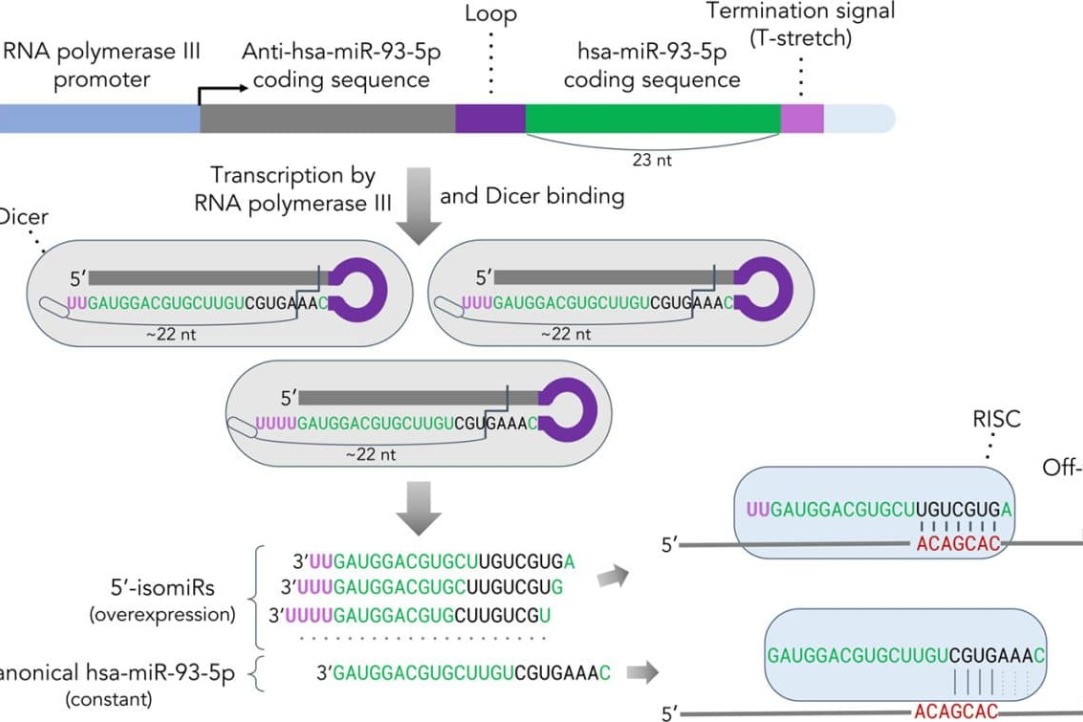
One commonly employed method for determining the function of a gene is to induce overexpression of the gene within the cell. This process also may involve the utilization of microRNAs, which are encoded by genes and produce short products that consist of only 20 nucleotides. In order to achieve this objective, artificial constructs containing the target gene are introduced into the cellular DNA. The accuracy of the results depends on the correct design of these constructs.This holds particular significance for microRNAs, as they play a regulatory role in the expression of other genes.
Employees of the International Laboratory of Microphysiological Systems together with colleagues from the Institute of Biological Chemistry of the Russian Academy of Sciences have found that expression of constructs carrying microRNA genes produces not a single product, but a set of forms with 5’-ends differing by several nucleotides. This occurs because RNA polymerase III incorporates 1-3 uracil nucleotides to the 3’-end of a construct introduced into the genome during expression. As a result, instead of a single transcript, a series of transcripts with differing lengths is produced. After slicing by Dicer RNase, these transcripts yield a set of microRNAs with different 5’-ends.These “additional” microRNAs recognize and regulate genes that are not subject to regulation by the target microRNA introduced into the genome, thus substantially distorting the study’s outcomes.
The detection of this heterogeneity can only be accomplished through sequencing, necessitating the development of additional design requirements for genetic constructions in order to mitigate the issue. The results have been published in the journal Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. Read morehere,hereandhere.